

CRI-MAP Tutorial - Data File Formatting
- 1. The initial data file,
chr#.gen
- Ex. 1 - create an input file
- 2. Auxilliary data files,
chr#.loc
chr#.ord
chr#.par
chr#.dat
(prepare option)
- Ex. 2 - prepare the auxilliary data files
- 3. Merging data files with common loci or families
(merge option)
- Ex. 3.1 - merge two chr#.gen files
- Ex. 3.2 - prepare new auxilliary data files
You must supply the initial data file to CRI-MAP. It is a plain
"ascii" text file that may be created and edited in almost any
word-processing programme. chr#.gen
data files always have the form:
(Section 1, with info for the entire pedigree collection:)
# of families
# of loci
locus_name1 ln2 ln3 etc. (NB: locus names have a 15 character limit)
(Section 2, with the pedigrees:)
(Section 2.1-f, with general info for the first family:)
family 1 id (any unique name or number designation)
# of family members
(Section 2.1-i, with the data for each individual from the first family:)
individual 1 id# mother's id# father's id# sex id#
locus_name1allele1# ln1a2# ln2a1# ln2a2# etc.
individual 2 id# mother's id# father's id# sex id#
ln1allele1# ln1a2# ln2a1# ln2a2# etc.
etc.
(Section 2.2-f, with general info for the second family:)
family 2 id
# of family members
(Section 2.2-i, with the data for each individual from the second family:)
individual 1 id# mother's id# father's id# sex id#
locus_name1allele1# ln1a2# ln2a1# ln2a2# etc.
individual 2 id# mother's id# father's id# sex id#
ln1allele1# ln1a2# ln2a1# ln2a2# etc.
etc.
and so on ...
The manual shows this short generic example.

Points to note:
- locus names are separated by spaces, so any particular locus name may NOT
contain a space character
- locus names and family names may be any combination of letters, punctuation,
and/or numbers
- "id#"s are integers - ALL alleles must be coded as
integers
- for individuals without parents in the pedigree, "mother's id#
& father's id#" are coded as 0
- for "sex id#", female=0 male=1 unknown=3
- unknown "allele#"s are coded as 0
- for X-linked loci, code the non-existent second alleles in males (for the
Y-chromosome) with an unused ("dummy") allele number
-
- Exercise CRI-MAP 1: create an
input chr#.gen file
- Using this key to map allele names to allele numbers, code the following pair of
pedigrees into an input file for CRI-MAP. Use a word processor
that allows file saving in
text format. Name the
file chr37.gen.
Chromosome 37 Loci
Locus Name allele (code #) allele (code #) allele (code #)
A a (1) A (2)
B b (1) B (2)
C c (1) C (2) c* (3)
D d (1) D (2) d* (3)
E e (1) E (2)
Family LMN.3 Chr. 37 Loci
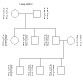
 Family OPQ.7 Chr. 37 Loci
Family OPQ.7 Chr. 37 Loci
NB: When using your own data, if they are already in the
correct format for analysis by the Linkage suite of programmes,
you may convert them to CRI-MAP chr#.gen
format with the LNKTOCRI utility. 
 2. The auxilliary data files - via the prepare option
2. The auxilliary data files - via the prepare option
- chr#.loc
- chr#.ord

- chr#.par

- chr#.dat

With the chr#.gen file written, you submit it
to the prepare option of CRI-MAP to create standard
versions of the auxilliary data files. ALL CRI-MAP
options are applied to datafiles with commands of this form:
prompt> crimap # option
For example, to perform "twopoint" linkage analysis on data for
chromosome 12 (you will already have datafiles named chr12.gen, chr12.dat,
chr12.loc, chr12.ord, & chr12.par), the command is:
prompt> crimap 12 twopoint
The programme then responds by displaying the results of the analysis on
the screen. Unlike most other CRI-MAP
options, however, prepare is an interactive one; before
it completes the preparation of the auxilliary datafiles, you must reply to a
series of questions, mostly having to do with accepting default values for
various parameters and filenames used in the analytical options.
-
- Exercise CRI-MAP 2: prepare the
auxiliary data files
- If you haven't already done so, copy your plain text
chr37.gen
file into a new directory in your filespace at your EMBnet Node
host computer. Name the directory Crimaptutl, and switch
to it when ready to proceed. (Unfamiliar with UNIX? Check the UNIX
Commands Short List page.)
- Enter the command crimap 37 prepare at the prompt, and
accept the default values or enter the answers shown
in the series of questions that follows.
Do you wish to enter any new haplotyped
systems?", enter n.
- When asked " Do you wish to hold any additional recombination
fractions fixed?", enter n.
- Choose build as the next option to run, by entering
"1".
- When asked "
Do you wish to build the map incorporating ALL loci
in decreasing order of their informativeness (the usual procedure)?,
enter y.
- And finally, when asked " OK to set up new parameter
file?, enter y.
NB: If you see the warning:
NONINHERITANCE: family "fam_name", individ id#, locus #
your chr37.gen file has an error!
Although we have chosen build as the next option to run, first examine
the structure and the content of the datafiles just created from the chromosome
37 pedigrees. These datafiles summarise different aspects of the data and
details of the subsequent processing by CRIMAP.
- chr37.loc
is a file holding only information on the loci.
- Quoted below is
the chr37.loc file derived from only Family LMN.3;
it shows the number of loci, the index numbers and names of each, plus two
columns for "informative meioses".
Genotype file chr37lmn.gen
Number of loci: 5
#inf. mei. #inf. mei.(phase known)
0 A 3 3
1 B 3 3
2 C 7 3
3 D 11 0
4 E 4 0
Traditionally, an informative meiosis is one that, firstly, leads
to offspring, and secondly, occurs in a parent who is (usually) at least
a double heterozygote. In Family
LMN.3, the grandfather is a triple heterozygote who produces four children.
Thus, he yields four informative meioses (one per child) for each of the
three loci: C, D, & E. His son who is a triple heterozygote (loci A,
B & C) produces three children, thus yielding three informative meioses
for each of these three loci. Summing up, the totals are: A-3, B-3, C-7,
D-4, & E-4. Why then has locus D eleven informative
meioses, according to CRI-MAP?
The prepare option has determined that both the grandmother and
the son's partner - even though they are only single heterozygotes - also
provide informative meioses for the D locus. There are four meioses in the
grandmother (her four children) plus three more from her son's partner.
This adds the extra seven informative meioses for locus D.
Note that the informative meioses from the son are also phase known.
This means that we know which set of five alleles for these five loci came from
each parent. We know the original allele complements (phase) of his two
chromosomes. He received a chromosome with alleles A b c* d E
from the grandmother, and another with alleles a B C d E from
the grandfather. Knowing the phase of the chromosomes reduces the number of
possible original-states the programme must explore in its analysis, thus
speeding result and increasing its accuracy.

- chr37.ord is
the file holding the initial information on locus order.
- Quoted below is the initial chr37.ord file produced by
prepare.
1
1 2
2 0
- For this simple chromosome 37 example, the prepare option
recognises only one set of possible orders, and this set has only one member,
the trivial order consisting of the two most informative loci. The order itself
(trivial because it has only two loci) is shown via the index numbers
of the loci: "2 0".
As the chromosome map is
estimated by CRI-MAPs build option, the
chr37.ord file will be modified and updated, finally reporting ALL
likely locus orders. A "likely" locus order is one within a given
range of the "most likely" locus order(s), the one(s) with the lowest
LOD score. We'll examine this file again after using the build option.

- chr37.par
contains the values for all the parametres
used by the various CRI-MAP options.
- The values for the many parametres are the default ones accepted during
the prepare operation. Detailed descriptions of the parametres, their
default values, and how they are used by the CRI-MAP options
are in the manual.
For now, notice that
the chr37.par file holds the names of the other auxilliary data files,
dat_file chr37.dat *
gen_file chr37.gen *
ord_file chr37.ord *
tells not to use the chr37.ord file in the subsequent
build option
use_ord_file 0 *
though it may be altered
write_ord_file 1 *
and shows the index numbers of the loci having an established order
ordered_loci 2 0 *
and of those waiting to be established.
inserted_loci 1 3 4 *

- chr37.dat shows
the phase known data available to the build option.
- The phase known information is organised by chromosome and by
family in this file, as opposed to by locus in chr37.loc.
 3. Merging data files with the merge option
3. Merging data files with the merge option
As you acquire new data, you may add it to existing data files automatically.
merge allows the addition of data when either loci or families are
shared between separate chr#.gen files.
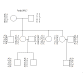
For example, we could increase the amount of phase known information for the
chromosome 37 dataset if we knew the genotypes of the parents of the two males
who "bred into" family
OPQ.7 .
-
- Exercise CRI-MAP 3.1: merge two
chr#.gen files
- Copy the plain text
chr37OPQ.gen
file into your Crimaptutl, directory.
- Enter the command crimap 37 merge at the prompt, and
enter the answers shown
in the series of questions that follows.
first input file =", enter chr37.gen.
- When asked " second input file =", enter chr37OPQ.gen.
- When asked " output file =,
enter chr37a.gen.
NB:You will see warnings similar to:
MISMATCH IN PEDIGREE 537728400, individual 7
MISMATCH IN PEDIGREE 537728400, individual 8
The chr37OPQ.gen file has no error - these two individuals are the two
males who have just had their parents added to the pedigree. As such, their
"mother's id#" & "father's id#"
values change from "0" (unknown) to one of the four new pedigree
members.
What additional information is now available to CRI-MAP? A
quick way to check is to compare the chr37.loc file with the new
chr37a.loc file you can create via the crimap 37a prepare
command.
-
- Exercise CRI-MAP 3.2: prepare new
auxilliary data files using the added data
- Enter the command
crimap 37a prepare at the prompt, and
accept the default values or enter the answers
as before.
Note that locus E is now reported as being more informative than locus D by the
prepare option output.
Comparison of chr37.loc with
chr37a.loc shows four more informative meioses
overall, 10 more being phase known, and three of these new phase known
meioses occurring at locus E.
Please continue with the build, twopoint, flipsn,
instant, & fixed options, in
Part 2 - Mapping & LOD scores










 Family OPQ.7 Chr. 37 Loci
Family OPQ.7 Chr. 37 Loci