Chromosome Linkage and Mapping
Sometimes, two traits are visible together. For instance, in corn, the traits for color and kernel fullness are usually carried on one gene, and the recessive alleles for these traits can be carried on an another. When both the recessive or the dominant alleles for two traits are on the same chromosome, it is called the cis phase. However, when a recessive and dominant allele for the different traits are on the same chromosome, we call it the trans phase.
So, when two parents are crossed continually, very seldom do you see a mixing of this pattern. This is called linkage. However, when colored, shrunken kernels apper less than the two homozygous types, we can assume that the two homologous chromosomes crossed over and caused a variation. This is called crossing over.
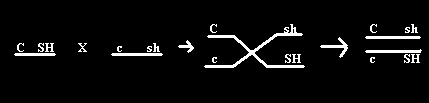
Normally, when a person crosses a heterozygous colored, full kernel corn plant to a homozygous recessive, we would expect to get:

The two types with the higher numbers represent the two parent-type gametes, since not all cells will crossover. The other two represent the single crossovers (SXO).
Since we have figured out that crossing over has occurred, we can map the distance between these two traits on the chromosome. The closer the traits are on the chromosome, the less likely crossing over will occur.
To figure map distance, we can look at the numbers to determine the parent crosses, and whether or not the genes are in trans or cis formation. Then we can use the equation:
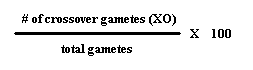
This percent can be used as the number of map units apart the two genes are. If the two genes are further than 50 units, crossing over will not be a factor.
To determine if crossing over exists, it is best to cross a heterozygous plant to a homozygous recessive plant. Then once can calculate the possible gametes from the heterozygous parent. Remember, the larger numbers of the results will show the parental genotypes.
For example:

If we know the map units between two traits, we can figure out the outcome of a cross of a heterozygous individual to a homozygous recessive. If two traits are 10 map units apart, we know that this also can be placed as a percent and represent the amount of crossovers occurring. Since there are two possible crossover gametes that occur 10% of the time, we can divide .10 by 2 to get .05 of a chance per crossover gamete. To figure the parental occurrence, we know that 100 - 10 = 90% occurrence, and that there are two possible gametes, so we get a .45 chance for each.
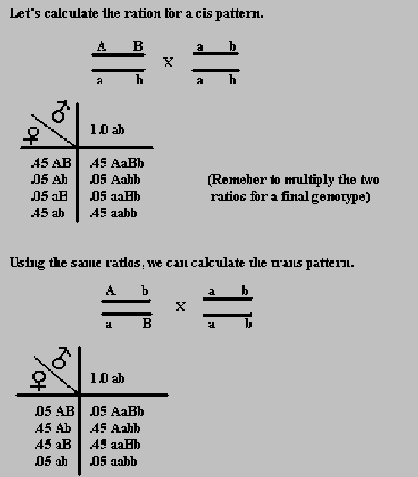
- Continue to Mapping Problem 1
- Return to Main Menu
|
|
© 2003-2026:
USA · USDA · NRPSP8 · Program to Accelerate Animal Genomics Applications.
|
||